python操作txt檔案中資料教程[3]-python讀取資料夾中所有txt檔案並將資料轉為csv檔案
阿新 • • 發佈:2018-11-26
python操作txt檔案中資料教程[3]-python讀取資料夾中所有txt檔案並將資料轉為csv檔案
覺得有用的話,歡迎一起討論相互學習~Follow Me
參考文獻
python操作txt檔案中資料教程[1]-使用python讀寫txt檔案
python操作txt檔案中資料教程[2]-python提取txt檔案
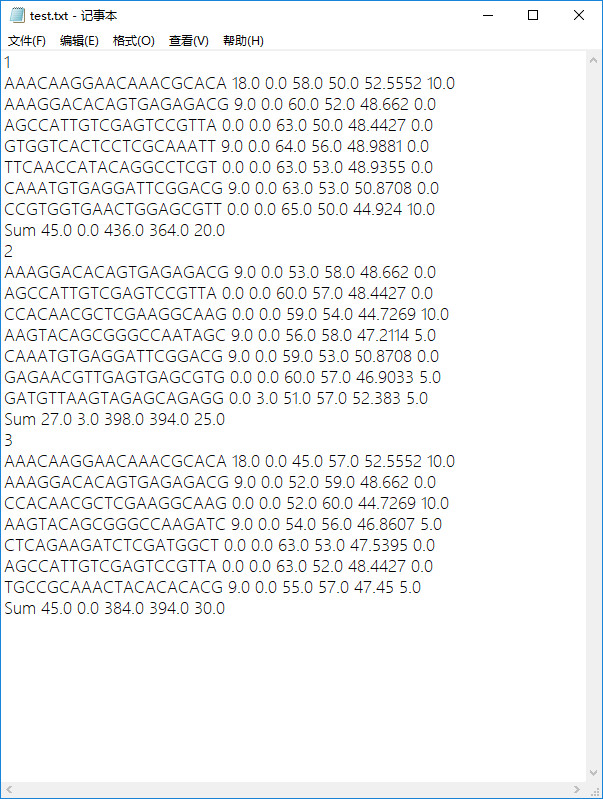
- 原始txt檔案

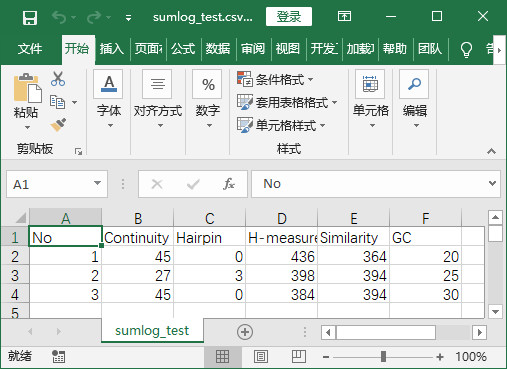
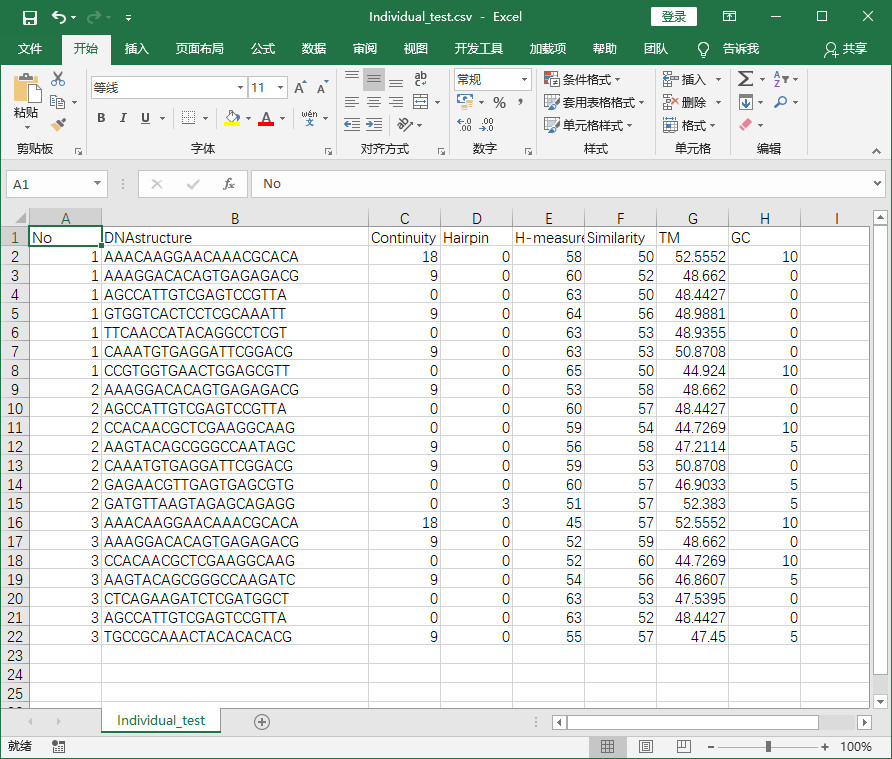
- 程式實現後結果

程式實現
import csv import os SUM_LOG_FILE = [] # sum_csv檔名 INDIVIDUAL_LOG_FILE = [] # individual_csv檔名 File_Name = [] # txt_檔名 DNA_Group = 7 # 表示每7條DNA組成一個組 Sum_log_file_header = ["No", "Continuity", "Hairpin", "H-measure", "Similarity", "GC"] Individual_log_file_header = ["No", "DNAstructure", "Continuity", "Hairpin", "H-measure", "Similarity", "TM", "GC"] def Read_Files(filename): DNA_log = [] # 精英種群個體日誌mod9=1-8 Sum_log = [] # 精英種群總體日誌mod9=0 sum_evaindex = [[] for i in range(6)] Individual_evaindex = [[] for i in range(8)] with open(filename, 'r') as f: i = 1 for line in f.readlines(): if i%9 == 0: Sum_log.append(line) else: DNA_log.append(line) i = i + 1 f.close() Sum_no = 1 dna_log_no = 0 for Sum in Sum_log: sum_eva_index = Sum.split("\n")[0].split(" ")[1:] sum_evaindex[0].append(int(Sum_no)) sum_evaindex[1].append(float(sum_eva_index[0])) # Con sum_evaindex[2].append(float(sum_eva_index[1])) # HP sum_evaindex[3].append(float(sum_eva_index[2])) # Hm sum_evaindex[4].append(float(sum_eva_index[3])) # Si sum_evaindex[5].append(float(sum_eva_index[4])) # GC Sum_no = Sum_no + 1 for dna_log in DNA_log: # 獲取序號值 if (dna_log_no + 1)%8 == 1: for i in range(DNA_Group): Individual_evaindex[0].append(int(dna_log.split("\n")[0])) else: # 獲取各項指標 Individual_evaindex[1].append(dna_log.split("\n")[0].split(" ")[0]) # 所有DNA序列全部記載,使用原有的str字串型別記載 Individual_evaindex[2].append(float(dna_log.split("\n")[0].split(" ")[1])) # DNA序列的連續值Con,注意要轉換為浮點數型別 Individual_evaindex[3].append(float(dna_log.split("\n")[0].split(" ")[2])) # Hp莖區匹配 Individual_evaindex[4].append(float(dna_log.split("\n")[0].split(" ")[3])) # H-measure Individual_evaindex[5].append(float(dna_log.split("\n")[0].split(" ")[4])) # Similarity Individual_evaindex[6].append(float(dna_log.split("\n")[0].split(" ")[5])) # TM Individual_evaindex[7].append(float(dna_log.split("\n")[0].split(" ")[6])) # GC dna_log_no = dna_log_no + 1 return sum_evaindex, Individual_evaindex # 將資料寫入csv日誌檔案中 def Write_SumFiles(filename, sum_evaindex): with open(filename, "w", newline='') as f: writer = csv.writer(f) writer.writerow(Sum_log_file_header) # 注意,此處使用writerow而不是使用writerows for i in range(sum_evaindex[0][-1]): writer.writerow( [sum_evaindex[0][i], sum_evaindex[1][i], sum_evaindex[2][i], sum_evaindex[3][i], sum_evaindex[4][i], sum_evaindex[5][i]]) f.close() def Write_IndividualFiles(filename, sum_evaindex, Individual_evaindex): with open(filename, "w", newline='') as f: writer = csv.writer(f) writer.writerow(Individual_log_file_header) # 注意,此處使用writerow而不是使用writerows for i in range(sum_evaindex[0][-1]*DNA_Group): writer.writerow( [Individual_evaindex[0][i], Individual_evaindex[1][i], Individual_evaindex[2][i], Individual_evaindex[3][i], Individual_evaindex[4][i], Individual_evaindex[5][i], Individual_evaindex[6][i], Individual_evaindex[7][i]]) f.close() def file_name(file_dir): for files in os.listdir(file_dir): if os.path.splitext(files)[1] == '.txt': File_Name.append(files) SUM_LOG_FILE.append("./test/sumlog_" + os.path.splitext(files)[0] + ".csv") INDIVIDUAL_LOG_FILE.append("./test/Individual_" + os.path.splitext(files)[0] + ".csv") # 獲取當前目錄下所有txt檔名 file_name(".") for i, j, k in zip(File_Name, SUM_LOG_FILE, INDIVIDUAL_LOG_FILE): print(i) print(j) print(k) Sum_Evaindex, Individual_Evaindex = Read_Files(i) Write_SumFiles(filename=j, sum_evaindex=Sum_Evaindex) Write_IndividualFiles(filename=k, sum_evaindex=Sum_Evaindex, Individual_evaindex=Individual_Evaindex)
